UH Biocomputation Group
Introduction
This purpose of this page is to document the steps required to update the UHBiocomputation website. The steps here apply only to users that have write access to the repository. There is a different SOP document for non admins. The website is generated using a Python tool called Pelican which converts the source files into HTML files with the required template design and so on. The sources are kept in the Github repository so that:
- They are backed up.
- Any errors can be easily rolled back.
- They are easily accessible to anyyou that may need to modify them.
- Non administrators that would like to make modifications can make them without the admins having to actively do all the work.
Since Pelican is written in Python, it can be installed on all operating systems. The recommended method is to use a Python virtual environment.
# change pelican-virt to wherever you want to set up the folder $ virtualenv -p /usr/bin/python3 pelican-virt #on Ubuntu $ virtual-3 pelican-virt #on Fedora
One can then install pelican and the other requirements in this virtual environment.
$ source pelican-virt/bin/activate #activate the virtual environment $ pip install pelican icalendar pybtex Pygments feedgenerator bs4 BeautifulSoup4 pytz $ deactivate #deactivate the virtual environment
More information on installing Pelican can be found here.
Git is required to get the sources. On Linux variants, Git is available using the default package managers.
$ sudo dnf install git-all #On Fedora $ sudo apt-get install git #On Ubuntu variants.
Git can also be installed on Windows and Mac systems using the installers available here. The default Git installation provides gitk, but there are other graphical clients available for install too.
To publish using FTP, the Makefile uses the lftp command. This can also be installed on Linux systems quite easily:
$ sudo dnf install lftp #On Fedora $ sudo apt-get install lftp #On Ubuntu variants.
Filezilla is an excellent graphical FTP application that can also be used.
There are three simple steps required to make updates to the website:
- Get the sources and make the changes.
- Preview, confirm and commit.
- Generate the final pages and push to hosting.
Get the sources and make the changes
The sources are plain text files which can be either written in markdown or restructured text using your favourite text editor on your favourite operating system. If you haven't already cloned the repository, you will need to do so:
$ git clone --recursive git@github.com:UHBiocomputation/UH-biocomputation-web.git
The recursive flag is required so that you also clone the pelican-plugins submodule. If you already have a clone of the repository, please make sure it is up to date with the latest commits (other people may have made commits too):
$ cd ~/website #wherever your clone resides $ git pull $ git submodule foreach git pull origin master $ git submodule foreach git checkout master
The directory structure of the sources is as follows:
[asinha@ankur UH-biocomputation-web]$ tree -d -L 2 . ├── content │ ├── files │ ├── images │ └── pages ├── output ├── pelican-plugins └── pelican-theme-gum ├── static └── templates
- All the source files are stored in the content directory. Posts in Pelican are divided into two categories: blogs and static pages. All the pages in the site header are static pages. These reside in content/pages. The posts that make up the "news" section are blog posts and go straight in the content directory. Various images and files that are linked to in either pages or posts go into their respective folders.
- The generated output files go in the output directory. The contents of this directory are then uploaded to our hosting webspace and constitute the actual website.
- The pelican-theme-gum directory holds the theme template and css files for the site. Again, unless tinkering with the theme, you need not touch this directory at all.
- The pelican-plugins directory is a git repository in itself. Most of the time, you will not deal with this directory at all. We only use the tag_cloud plugin. The commands mentioned enough will correctly clone this directory.
Once you've cloned the repository, make your changes and save them. Note: Changes can be made to files using the Github web interface also. However, to run pelican, preview changes, and then generate the final website, you have to download the sources anyway.
Adding a new post
The Makefile includes a helper command to create a new post for the website:
make newpost NAME="Title of post" EDITOR="name of editor one uses: gvim, gedit"
This generates a new template file in the content folder that can be
modified. If the EDITOR environment variable is defined, it will also
open this template file in the specified editor:
$ make newpost NAME="A test post" EDITOR="gedit" ... messsages regarding setting up new template post ... gvim -v /home/asinha/Documents/02_Code/00_repos/01_others/UHBiocomputation/UH-biocomputation-web/content/20180125-a-test-post.rst
When the post has been completed, one can save it, and preview-publish as documented below.
Adding a new seminar
The Makefile includes a helper command to create a new seminar for the journal club:
make newseminar AUTHOR="Full name of presenter" NAME="Title of the talk"
In order for this to work, one must first generate the seminar_file.bib and save it in the scripts folder.
The seminar_file.bib should contain all papers relative to that seminar. An easy way to generate the seminar_file.bib is by saving the papers to Zotero, then exporting them in .bib format.
When the post has been completed, one can save it, and preview-publish as
documented below.
Remeber, once created a new seminar, to push your commits on GitHub with this message:
JC on YYYY-MM-DD by speaker
New seminar - special cases
One may need further instructions for these special cases:
The name of the seminar is different from the title of the paper/s to present. In this case, the key to the main paper must be provided as so:
make newseminar AUTHOR="Full name of presenter" NAME="Title of the talk" KEY="Main_paper_key"
Paper keys can be found in the
seminar_file.bib.The presenter does not reference any paper. In this case, generate a dummy
seminar_file.bibwhich must contain the following information:- Title: title of the presentation
- Author: name of the presenter
- Journal: "dummy"
- Abstract: summary of the presentation
- DOI: link to the Biocomputation group seminar page
- Date: year of the presentation
Then procede creating the new seminar as described previously and remove the reference generated by the dummy
seminar_file.bibfrom the new seminar.
Updating the journal club rota
The rota is mangaged using a CSV file in the scripts folder. The
current file is rota-2025.csv. Each line in this file represents an
entry that must be added to the ical file and the seminar page
05-seminars.rst. The seminar page is set up to source the generated
rota page. The Makefile takes care of generating the rota and ical files, and
copying them to the required locations.
Setting a speaker for the JC session requires 2 steps: (1) generating a file
with the abstract of the talk and (2) modifying the CSV file in the
scripts folder. Generation of the file with abstract (1) is done by
adding a new seminar which is described above. The CSV file should be appended with the information about
the talk and should be done according to following formula:
Name of presenter,"Title of talk","Location of rst post in contents folder",Date of event(YYYY-MM-DD),Start time in 24h format,End time in 24h format,Location(0 represents default, online),Whether or not this entry should be added to the seminars page: 1 = Yes, 2 = No
It is possible to add a speaker without title and abstract file (leave the both sets of quotation marks empty). The abstract file name is the name of the post file created in (1). An example of fully modified formula is presented below:
Ankur Sinha,"Associative properties of structural plasticity based on firing rate homeostasis in a balanced recurrent network of spiking neurons","20170904-associative-properties-of-structural-plasticity-based-on-firing-rate-homeostasis-in-a-balanced-recurrent-network-of-spiking-neurons.rst",2017-09-08,1600,1700,0,1
Note that when a new rota is started (at the beginning of September and January),
there are 3 steps to be done. First, a new CSV file should be added
to the scripts folder (as described above). Second, the newly
created CSV file needs to be sourced in the content/pages/05-seminars.rst
file. The following example shows what text should be added just below the first
paragraph:
September 2021 - ---------------- .. include:: rota-2021b.txt
Third, changing the starting date for the rota. This is done in the
file scripts/generate_rota.py, under line 72:
# ===-===-===-===-===-===-===-===-===-===-===-===- # TODO this should be changed to be loaded from the rota csv files self.start_date = date(2023, 1, 27) # ===-===-===-===-===-===-===-===-===-===-===-===-
If an entry should be added to the ical file, but not to the seminar page, for example, a colloquium talk that will not have a corresponding abstract published on the website and should not be listed on the website seminar list, one can set the last field to 0.
Once updated, the ical files and updated rota for the website can be generated using:
make rota
Note that make html is set up to run make rota already.
Preview, confirm, and commit
The Pelican Makefile has the commands required to preview and publish the website.
Once the required changes have been made and the file saved, preview the website to confirm that everything works as expected. To do this, in the main directory where the Makefile resides, run:
$ make html Pelican /home/asinha/Documents/02_Code/00_repos/others/UHBiocomputation/UH-biocomputation-web/content -o /home/asinha/Documents/02_Code/00_repos/others/UHBiocomputation/UH-biocomputation-web/output -s /home/asinha/Documents/02_Code/00_repos/others/UHBiocomputation/UH-biocomputation-web/pelicanconf.py Dyou: Processed 4 articles, 0 drafts, 5 pages and 7 hidden pages in 1.04 seconds.
If this command completes without errors, preview the website:
$ make serve
This sets up a local server that serves the website at localhost:8000 so that you can preview your changes. To stop this server, hit Ctrl + C. If everything is OK, you can commit your changes:
$ git add . $ git commit -m "A sensible commit message." $ git push -u origin master
Generate the final pages and push to hosting.
The last step is to generate the final version and upload the site to the webhosting location. If ftp access is available, this can be dyou with you command:
$ make ftp_upload #runs make publish for you
The Makefile already contains the required details. The command will ask you for the ftp password and do the rest. When it finishes uploading, your site should be up to date.
Merging pull requests
Non admins can open pull requests as documented in the nonadmin SOP document. Admins only need to review the proposed changes, merge them if they're OK and then republish the website. This saves the admins from doing all the writing work required to update the website.
Pull requests that have been correctly created do not require anything other than a button click. They will specify that the pull request was made correctly and that the merge can be made without issues as shown in the figure below:
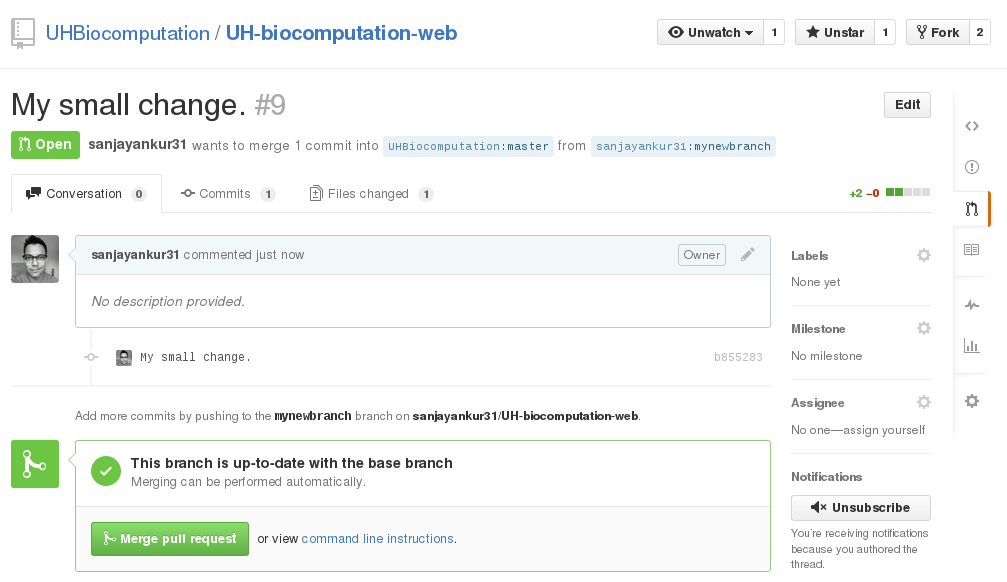
However, an admin should generally check that the changes made in the pull request are all correct. For small changes, you can just click on the "Files changed" tab and verify the changes. If they're OK, you can merge the pull request right away. For larger changes, you will have to checkout the person's branch, test the changes and then merge the request. The instructions to do this can be seen by clicking the "command line instructions" link in the merge ticket.
Once you've merged the pull request, the copy of the repository on your local system will need to be updated, the site regenerated, and uploaded. This should generally be as simple as:
$ cd website-directory $ git pull origin master #this will pull the latest commits that you or others have merged or made $ make html; make serve #Again check that everything is OK by going to localhost:8000 $ make ftp_upload #If everything is correct, upload the lastest version.
That should be it!