UH Biocomputation Group
Research in the Biocomputation Laboratory involves the application of computational methods to biological problems, and the development of computational applications based on inspiration from biology. The research is coordinated by Prof. Volker Steuber and falls into the subject areas Computational Neuroscience, Machine Learning, Computational Systems Biology, Computational Algebraic Biology, and Bioinformatics.
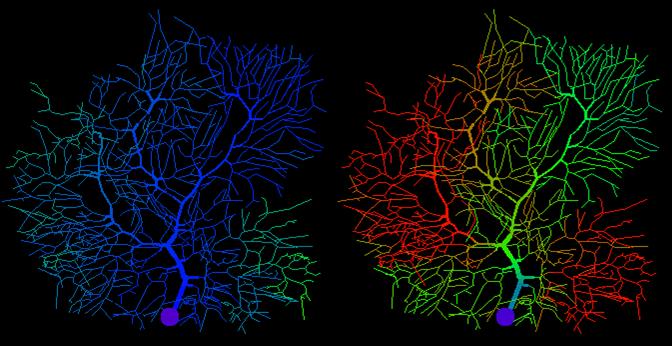
Research in Computational Neuroscience focuses on the development, simulation and analysis of computational models of neurons and neuronal networks in order to understand information processing in the brain. To constrain and test the models, the Computational Neuroscience Team collaborates closely with world-leading neuroscientists in the UK, France, Japan, Israel, the Netherlands and the USA. The team applies a multi-scale systems biology approach, using models at scales ranging from biochemical processes to neuronal networks and behaviour, and interacting continuously with anatomists, neurophysiologists and biochemists that investigate neuronal systems at multiple levels of description.
Research in Brain-inspired Data Science and Neuromorphic Computing translates processes from Neuroscience to Computing, aiming to push the envelope in our Data Science efforts. We develop new algorithms for data analysis on Neuromorphic Hardware, i.e. silicon chips tailor-made to operate in a brain-like fashion, based on a non-von-Neumann architecture. In designing those algorithms we take inspiration from biological neuronal networks involved in sensory computation in order to exploit this radically new way of computing. We apply these techniques alongside conventional Machine Learning approaches for data analysis for life science, Olfaction and Chemical sensing, and combine them with robotics applications within the emerging research field of Neurorobotics.
Research in Computational Systems Biology involves the development of visualization and mixed-mode stochastic simulation tools for biochemical reaction networks. Current and potential applications of these tools include basic medical research such as studying the effect of anti-cancer drugs, and the visualization of biological processes for education purposes (in collaboration with the School of Creative Arts at UH).
Generating impact outside academia is part of the strategy of the Machine Learning Team. The team applies biologically inspired Machine Learning techniques to a large range of real-world problems including the analysis of bit error correction in network transmissions, software defect detection and the prediction of trans-dermal drug penetration (in collaboration with MedPharm). Members of the Machine Learning Team also contribute to all other areas of Biocomputation Research.
Research on Computational Algebraic Biology brings the methods of finite group and Krohn-Rhodes automata theory to the problems of detecting hierarchical structure, invariant subsystems, and local control structure within systems biology and related areas andhas been applied to biochemical and genetic regulatory systems (such as the p53-mdm network in cancer). It also serves as a basis for developing Interaction as a unconventional computation paradigm (FP7 BIOMICS project) with security and distributed computing applications.
Another area that impacts on medicine is the Bioinformatics. The Bioinformatics Team uses computational techniques to study regulatory regions in DNA and their role in complex diseases such as type 1 diabetes.
We are also involved in collaborative initiatives that aim to develop Spiking Neural Networks for the Control of Simulated Agents.